This notebook provides an introduction to Python Geospatial Libraries, mainly GeoPandas (including aspects of Pandas), and Shapely. This notebook was created by Becky Vandewalle based off of prior work by Dandong Yin.
Introduction¶
Pandas is a Python library for data analysis, featuring easy IO, powerful indexing, database operations, and categorical data support. Two primary data structures in Pandas are `Series` and `DataFrame`.
GeoPandas extends Pandas by adding core geographical functionality. You can do spatial database-like operations with this library.
Shapely is useful for working with vector data using set-like operators.
Useful documentation for these libraries can be found here:
Pandas https://pandas.pydata.org/pandas-docs/stable/
GeoPandas http://geopandas.org/
Shapely https://shapely.readthedocs.io/en/stable/manual.htmlMore information about DataFrames https://pandas.pydata.org/pandas-docs/version/0.22/dsintro.html#DataFrame
Shapely attributes and methods https://shapely.readthedocs.io/en/stable/genindex.html
# import required libraries
%matplotlib inline
import os
import geopandas as gpd
import pandas as pd
import numpy as np
import shapely
Import Data¶
The main dataset used in this notebook is stored as a dBase database file (.dbf). Pandas can not import a dBase file directly, so Geopandas is used to convert it to a format Pandas can read.
# import towns database
towns_temp_gpd = gpd.read_file(os.path.join('./pyintro_resources/data','towns.dbf'))
towns_table = pd.DataFrame(towns_temp_gpd)[:10]
countries = gpd.read_file(os.path.join(r'zip://pyintro_resources/data/ne_10m_admin_0_countries.zip'))
cities = gpd.read_file(os.path.join(r'zip://pyintro_resources/data/ne_10m_populated_places.zip'))
rivers = gpd.read_file(os.path.join(r'zip://pyintro_resources/data/ne_10m_rivers_lake_centerlines.zip'))
# check type
type(towns_table)
Lets check to make sure the data were properly loaded by looking at the first few rows containing town information (more on the head and tail functions later).
# show initial values of data frame
towns_table.head()
We will return to the towns database later. For now, lets start by exploring a Series.
Introducing the Series¶
A Series in Pandas is a one-dimensional labeled array capable of holding a wide variety of data. This array also has an index, which is like a label for each of the values in the array. The basic method to create a Series is to call:
pd.Series(data, index=index)
Here, data can be many different things, such as:
- a Python dictionary
- a numpy array
- a scalar value (like
5)
Anything like an array, iterable, a dictionary, or a scalar value, will work for data contents!
Basically, a Series is a list with an index!
Here are some examples of constructing a `Series`:
# create a basic series
pd.Series([0,1,2], index=['a', 'b', 'c'])
Note that index values are on the left.
# create a series using a dictionary of values and index values
pd.Series({'a' : 0, 'b' : 1, 'c' : 2})
# scalar values will be repeated to be the length of the index
pd.Series(5, index=['a', 'b', 'c'])
# but the opposite does not work
# this cell will fail to run!
pd.Series([5, 5, 5], index=['a'])
# if no index is provided, data values will be used as an index
pd.Series([0,1,2])
Once you have a series object you can access the data and index values directly:
# see type of series
myseries = pd.Series({'a' : 0, 'b' : 1, 'c' : 2})
type(myseries)
# list series values
list(myseries.values)
# list series index values
list(myseries.index)
Array-like Behavior¶
You can access elements of a `Series` using the index operator, just like for regular lists and tuples in Python. Remember that Python indices start with 0.
Here are a few examples:
# return the first element
myseries[0]
# select everything up until element index 2 (the third element)
myseries[:2]
# find the median value
myseries.median()
Operations can be performed element-wise for the series.
# see which elements are greater than the medium
myseries > myseries.median()
Use any or all to check if a comparison is true for the series as a whole.
# are any elements greater than the medium value?
any(myseries > myseries.median())
# are all elements greater than the medium value?
all(myseries > myseries.median())
You can also select values using a logical index series.
# select by greater than median
myseries[myseries > myseries.median()]
myseries
You can also pass a list of indices to select series elements out of order.
# select by list of indices
myseries[[2, 1, 1]]
Other operations are performed element-wise too. For example:
# add series values to self
myseries + myseries
# raise to the 3rd power
myseries ** 3
Dictionary-like Behavior¶
Since a `Series` has an index, you can also work with a Series as if it were a Python dictionary:
# find value with index 'a'
myseries['a']
# set value of 'd'
myseries['d'] = 5
myseries
# see if index value in series
'f' in myseries
# see if index value in series
'c' in myseries
'Naming' a series
A Series has an optional `name` parameter:
# see current name (not assigned)
print (myseries.name)
If you name a Series, the name will show up in the information at the bottom:
# set name
myseries.rename('stock_price')
Introducing the DataFrame¶
What happens when a Series gains a dimension? It becomes a `DataFrame`!
We can use the towns database we imported at the beginning of this Notebook to see how to check out a DataFrame. The head function is useful to see the first few rows of a DataFrame.
# see start of DataFrame
towns_table.head()
Similarly, the tail function can be used to check the end:
# see end of DataFrame
towns_table.tail()
Like a Series, a DataFrame has an index. This one is a numeric range.
# see index
towns_table.index
You can also list the DataFrame column names:
# see column names
towns_table.columns
And it is often useful to dig into its values:
# see values
towns_table.values
It can be useful to double check the size of the values array. We know from the index that there are 351 rows, and now we can see there are 17 columns (by a few different methods).
# see the size of the values array
towns_table.values.shape
# count columns
print (len(towns_table.values[0]))
print (len(towns_table.columns))
Using index selection we can see all values in one row.
# see one row
towns_table.values[5]
We can use the double index method to find a value within a particular and row and column:
# find specific value
towns_table.values[5][5]
Sometimes it is useful to know summary information about a DataFrame. The describe makes this clear:
# summarize DataFrame
towns_table.describe()
Transposing a DataFrame can also come in handy:
# transpose DataFrame
towns_table.T
It is also possible to sort a DataFrame by values in a particular column:
# sort DataFrame by column
towns_table.sort_values(by='POP2010')
From Series to DataFrame¶
Now that we are slightly more familiar with the DataFrame, we can go into more detail. Technically, a DataFrame is implemented as a list of named Series all with the same index values.
# construct a DataFrame
myDataFrame = pd.DataFrame(
{'one' : pd.Series([1., 2., 3.], index=['a', 'b', 'c']),
'two' : pd.Series([1., 2., 3., 4.], index=['a', 'b', 'c', 'd'])})
myDataFrame
Remember the name attribute for a Series? Since each column in a DataFrame is a Series, each column name is equivalent to the name of the Series for that column.
# show DataFrame column names
myDataFrame.columns
Manipulating a DataFrame¶
Columns in a DataFrame can be freely selected, added, deleted (as with the "dictionary" aspect of Series):
# create new column from existing columns
myDataFrame['three'] = myDataFrame['one'] + myDataFrame['two']
myDataFrame
Columns in a DataFrame do not have to all be the same data type (as we saw with the towns database):
# create new column from comparison
myDataFrame['flag'] = myDataFrame['one'] > 2
myDataFrame
It is simple to delete a column:
# delete column
del myDataFrame['two']
myDataFrame
Scalar values assigned to a column repeat to fit the length of the DataFrame.
# add column
myDataFrame['four'] = 4
myDataFrame
You can also add a new row to the DataFrame using the append function.
# add row to DataFrame
myDataFrame.append({'one':3,'three':2.0,'flag':False},ignore_index=True)
Note that the changes won't affect the original DataFrame unless you re-assign it.
myDataFrame
# add row to DataFrame
myDataFrame2 = myDataFrame.append({'one':3,'three':2.0,'flag':False},ignore_index=True)
myDataFrame2
Don't add rows dynamically too often, as append makes a full copy of the data, so constantly using this function can create a significant performance hit. A better way is to include all rows while creating the DataFrame.
You can also drop rows and columns:
# drop rows
myDataFrame.drop(['a','c'])
axis specifies whether you are referring to rows or columns:
# drop columns
myDataFrame.drop(['one','four'], axis=1)
Selecting Data in a DataFrame¶
Selecting data is a common task when dealing with a DataFrames.
Select by Label¶
As seen before, we can select data using label values.
# select column by index
towns_table['POP2010']
We can select just a few rows with index slicing:
# select rows
towns_table[0:3]
We can combine row selection using indices and selecting columns using labels using the `loc` function:
# select rows and columns
towns_table.loc[0:2,['POP2010','TOWN']]
# select row and columns
towns_table.loc[3,['POP2010','TOWN','POP1990']]
# select specific value
towns_table.loc[3,'POP2010']
Select by (only) Position¶
Column label values are not necessary - you can select values using just indices with the `iloc` function. Here are some examples:
# select row
towns_table.iloc[3]
# select some columns from row
towns_table.iloc[3,0:5]
# select some columns from all rows
towns_table.iloc[:,0:5]
# select rows and columns
towns_table.iloc[:2,:2]
Select by Condition¶
We can use conditional operators to select DataFrame slices that meet specific conditions. Here are examples:
# select by condition
towns_table['POP2010']>16000
# select by pop difference
(towns_table['POP2010']-towns_table['POP1990'])>100
# select by condition and pop difference
(towns_table['POP2010']>16000) & ((towns_table['POP2010']-towns_table['POP1990'])>100)
# select by condition or pop difference
(towns_table['POP2010']>16000) | ((towns_table['POP2010']-towns_table['POP1990'])>100)
# select values by condition
towns_table[towns_table['POP2010']>16000]
# select values by pop difference
towns_table[towns_table['POP1990']-towns_table['POP1980']<100]
# select values by pop difference and condition
towns_table[((towns_table['POP2010']-towns_table['POP1990'])>100) & (towns_table['POP2010']>16000)]
# create new database from selection
simp_db = towns_table.loc[:,['TOWN','POP1980','POP1990','POP2000','POP2010']]
simp_db
Simple Plotting¶
It can be quick to get a simple DataFrame plot up, but finessing the plot can take some time. Our first plot isn't that helpful...
# basic plot
simp_db.plot();
# transpose
simp_db.T
# this doesn't work!
simp_db.T.plot()
Lets try to get the town names to plot!
# rename columns with town names
simp_db2 = simp_db.T.rename(columns=simp_db.T.iloc[0])
simp_db2
# now drop town column
simp_db3 = simp_db2.drop(['TOWN'])
Now we have town names on our plot. Perhaps, a different chart type would work better though!
# plot
simp_db3.plot(figsize=(14,12));
# bar plot
simp_db3.plot(kind='bar',figsize=(14,12));
Introducing the GeoDataFrame¶
So far we have been working with Pandas' DataFrame. Geopandas' `GeoDataFrame` extends the DataFrame with very useful spatial functions.
Just as a reminder, we have been working with a DataFrame.
# a DataFrame
type(towns_table)
# start of table
towns_table.head()
We can easily convert the DataFrame to a GeoDataFrame:
# convert to GeoDataFrame
towns_gdb = gpd.GeoDataFrame(towns_table, geometry='geometry')
Do you notice anything different in the table?
# start of GeoDataFrame
towns_gdb.head()
Not much, but the type is different:
# a GeoDataFrame
type(towns_gdb)
What is so cool about a `GeoDataFrame`? For one thing, normal DataFrame data operations all still apply. However, simple plotting produces entirely different results!
# plot GeoDataFrame
towns_gdb.plot();
You can create a GeoDataFrame directly from a file:
# create GeoDataFrame from file
chicago_gdf = gpd.GeoDataFrame.from_file(os.path.join('./pyintro_resources/data',
'Chicago_Community.geojson'))
And plot it:
# plot GeoDataFrame
chicago_gdf.plot();
And start to make more detailed plots, like these below:
# plot GeoDataFrame column
chicago_gdf.plot(column='community', figsize=(14,12));
# plot GeoDataFrame
towns_gdb.plot(column='POP2010', cmap='hot_r', legend=True);
Hex Values in the Geography Column¶
Sometimes when you import geospatial data the geometry values may look a little off, like long strings of numbers and letters. If this is the case, it is likely hex-encoded.
# example of hex encoded geometry
import binascii
hex_geo = binascii.hexlify(towns_gdb.loc[0, 'geometry'].wkb)
hex_geo
#hex_geo = towns_gdb.loc[0, 'geometry'].wkb.encode('hex')
#hex_geo
If that is the case, you can use Shapely's wkb.loads function to convert it to something more readable (and workable).
# convert hex to polygon
non_hex_geo = shapely.wkb.loads(hex_geo.decode("ascii"), hex=True)
# look at polygon string
str(non_hex_geo)
# see polygon type
type(non_hex_geo)
This provides a nice opportunity to introduce Shapely!
Introducing Shapely¶
Shapely is a really helpful library for working with geometries. You can use it both for simple geometric operations and working with complicated real-world geometries.
Basic Shapes¶
Some useful shapes are Points, Lines, and Polygons. You can combine them into MultiPoints, MultiLineStrings, and MultiPolygons. Here are a few examples of creating and viewing basic shapes:
# create point
sh_point = shapely.geometry.Point(0, 1)
# view point
sh_point
# view object type
type(sh_point)
# create line
sh_line = shapely.geometry.LineString([(0, 0), (1, 1)])
# view line
sh_line
# view object type
type(sh_line)
# create polygons
sh_polygon1 = shapely.geometry.Polygon([(0, 0), (1, 1), (1, 0)])
sh_polygon2 = shapely.geometry.Polygon([(2, 2), (3, 3), (3, 2), (2.75, 2.25)])
# view polygon
sh_polygon1
# view object type
type(sh_polygon1)
# create multipolygons
sh_polygons = shapely.geometry.MultiPolygon([sh_polygon1, sh_polygon2])
# view multipolygons
sh_polygons
# view object type
type(sh_polygons)
Note that the length is two - there are two polygons!
# view length
len(sh_polygons)
Well Known Text, or `wkt` is a human-readable way to represent geometry.
# view text
sh_polygons.wkt
Geometric Attributes and Methods¶
Once you have constructed your shapes, there are a variety of attributes you can look up or methods you can use on the geometric objects. Here are a few examples:
# find centroid
sh_polygons.centroid
# find centroid text
sh_polygons.centroid.wkt
# find area
sh_polygons.area
# find length
sh_polygons.length
# find boundary
sh_polygons.boundary
# find bounds
sh_polygons.bounds
# find envelope
sh_polygons.envelope
A polygon is created from a set of rings representing interior and exterior coordinates.
# create polygon with hole
sh_polygon3 = shapely.geometry.Polygon([(0, 0), (0, 1), (1, 1), (1, 0)],
[[(0.25, 0.25), (0.75, 0.25), (0.75, 0.75)],
[(0.25, 0.5), (0.25, 0.75), (0.5, 0.75)]])
# view polygon
sh_polygon3
# view exterior coordinates
list(sh_polygon3.exterior.coords)
# view interior coordinates
for inter in list(sh_polygon3.interiors):
print (inter.bounds)
You can also simplify geometry:
# select complex geometry
or_geo = towns_gdb.loc[1, 'geometry']
or_geo
# simplify geometry
or_geo.simplify(50)
# simplify geometry further
or_geo.simplify(500)
Exploring Spatial Relationships with Shapely¶
(Materials adapted from https://github.com/jorisvandenbossche/geopandas-tutorial)
Spatial relationships describe how two spatial objects relate to each other (e.g. overlap, intersect, contain...).
The topological, set-theoretic relationships in GIS are typically based on the DE-9IM model. See https://en.wikipedia.org/wiki/Spatial_relation for more information.
Here are some spatial relations illustrated:
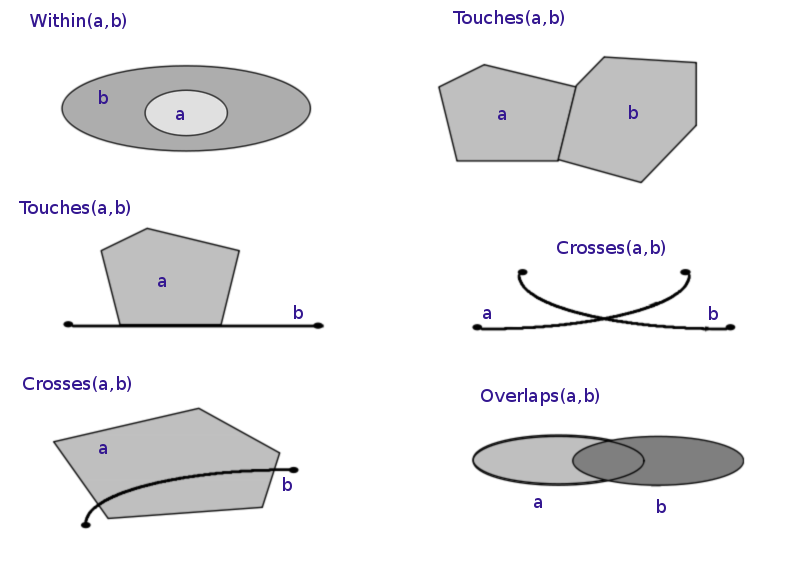
Lets explore these with Shapely!
We'll first set up some places and then see how their geometries interact.
# retrieve Belgium geometry from countries database
belgium = countries.loc[countries['NAME'] == 'Belgium', 'geometry'].unary_union
belgium
# retrieve Paris geometry from cities database
paris = cities.loc[cities['NAME'] == 'Paris', 'geometry'].unary_union
paris
# retrieve Brussels geometry from cities database
brussels = cities.loc[cities['NAME'] == 'Brussels', 'geometry'].unary_union
brussels
Let's draw a line between Paris and Brussels:
# create line between cities
city_connect = shapely.geometry.LineString([paris, brussels])
Now we can plot everything:
# plot objects
gpd.GeoSeries([belgium, paris, brussels, city_connect]).plot(cmap='tab10');
We can easily test for geometric relations between objects, like in these examples below:
# check within
brussels.within(belgium)
# check contains
belgium.contains(brussels)
# check within
paris.within(belgium)
# check contains
belgium.contains(paris)
Similarly, a GeoDataFrame (and GeoSeries) will also take spatial relationships:
# find paris in countries database
countries[countries.contains(paris)]
# select Amazon river from rivers database
amazon = rivers[rivers['name'] == 'Amazonas'].geometry.unary_union
amazon
# find countries that intersect Amazon river
countries[countries.intersects(amazon)]
# map countries that intersect Amazon
countries[countries.intersects(amazon)]['geometry']\
.append(gpd.GeoSeries(amazon)).plot(cmap='tab10');
Here is another example with the Danube:
# find and plot countries that intersect Danube river
danube = rivers[rivers['name'] == 'Danube'].geometry.unary_union
countries[countries.intersects(danube)]['geometry']\
.append(gpd.GeoSeries(danube),ignore_index=True).plot(cmap='tab10');
Performing Spatial Operations¶
Finally, we can perform spatial operations using Shapely. Let's look at a few examples:
# view amazon river
amazon
Shapely makes it easy to create a buffer around a feature:
# create a buffer
super_amazon = amazon.buffer(10)
super_amazon
# select Brazil from countries database
brazil = countries[countries['NAME']=='Brazil']['geometry'].unary_union
# plot Brazil, the Amazon river, and the buffer
gpd.GeoSeries([brazil, amazon, super_amazon]).plot(alpha=0.5, cmap='tab10');
Now we can view spatial relations between these polygons:
# intersects
super_amazon.intersection(brazil)
# union
super_amazon.union(brazil)
# union
brazil.union(super_amazon)
Note that for some spatial relations, the order in which they are called matters.
# difference
super_amazon.difference(brazil)
# difference
brazil.difference(super_amazon)
Lets end with a few more examples.
# find countries in South America
countries[countries['CONTINENT'] == 'South America']
# find geometry for countries
countries[countries['CONTINENT'] == 'South America'].unary_union
# plot all countries
countries.plot(column='NAME', cmap='tab20c', figsize=(14,14), categorical=True);
Now go forth and explore!